HWE and LD
Hardy-Weinberg equilibrium and Linkage disequilibrium are two basic concepts in population genetics.
Defination
- Hardy-Weinberg Equilibrium (HWE): allele and genotype frequencies in a population will remain constant from generation to generation in the absence of other evolutionary influences.
- Linkage Disequilibrium (LD): the non-random association of alleles at different loci in a given population.
So HWE is about the allele and genotype frequencies in one locus, but LD is about alleles at different loci.
Hardy-Weinberg Equilibrium
From Genotype to Allele frequencies
For $n_{AA}, n_{Aa}, n_{aa}$ is the count of genotype AA, Aa and aa, respectively. And $n = n_{AA} + n_{Aa} + n_{aa}$.
So, the count of A and a alleles are:
\[n_{A} = 2 \times n_{AA} + n_{Aa}\] \[n_{a} = 2 \times n_{aa} + n_{Aa}\]The frequencies of A and a alleles are:
\[p_{A} = \frac{n_{A}}{2n} = \frac{2 \times n_{AA} + n_{Aa}}{2n} = p_{AA} + \frac{1}{2}p_{Aa}\] \[p_{a} = \frac{n_{a}}{2n} = \frac{2 \times n_{aa} + n_{Aa}}{2n} = p_{aa} + \frac{1}{2}p_{Aa}\] \[p_{A} = 1 - p_{a}\]From Allele to Genotype frequencies - HWE
We cannot directly get genotype frequencies from allele frequencies. But under Hardy-Weiberge Equilibrium, we can do it by
\[p_{AA} = p_{A}^{2}\] \[p_{Aa} = 2p_{A}p_{a}\] \[p_{aa} = p_{a}^{2}\]The assumptions for HWE includes:
- random mating
- infinite population size (no genetic drift)
- no mutation
- no selection
- no migration
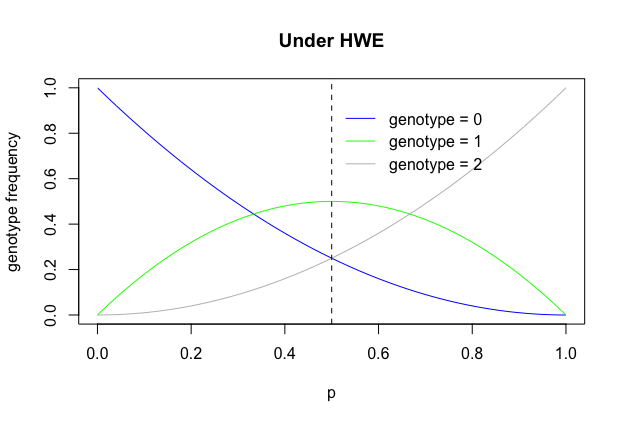
In addition, we can see genotype frequencies is a binomial distribution. If SNP genotpes are coded X = 0, 1 and 2 (alleles) and the allele frequency is p, then:
\[X \sim B(2, p)\text{ with }E(X) = 2p\text{ and }Var(X)=2p(1-p)\]We can plot the distribution of three different genotypes. In real genetics, we usually use the minor allele, so p is less than 0.5.
p = seq(0,1,0.01)
plot(p, (1-p)^2, ylab = "genotype frequency", main="Under HWE",type="l", col="blue")
lines(p, 2*p*(1-p), col="green")
lines(p, p^2, col="grey")
legend(0.5,0.95,"genotype = 0",lty = 1, col ="blue", bty="n")
legend(0.5,0.85,"genotype = 1",lty = 1, col ="green", bty = "n")
legend(0.5,0.75,"genotype = 2",lty = 1, col ="grey", bty = "n")
abline(v = 0.5, lty=2)
Test for HWE
In the GWAS QC step, we usually remove SNPs with extensive deviation from HWE because this can be indicative of a genotyping or genotype-calling error.
The two common ways to test HWE are Chi-square test and exact test.
For Chi-square test:
\[\chi^{2} = \frac{(n_{AA}-e{AA})^2}{e_{AA}} + \frac{(n_{Aa}-e{Aa})^2}{e_{Aa}} + \frac{(n_{aa}-e{aa})^2}{e_{aa}}\]where $e_{AA} = np_{A}^2$, $e_{Aa} = 2np_{A}p_{a}$, $e_{aa} = np_{a}^2$ and df = 1 (#genotype - #allele).
For exact test:
\[P(N_{Aa}|N_{A}) = \frac{n_{A}!n_{a}!n!2^{n_{Aa}}}{\frac{1}{2}(n_{A}-n_{Aa})!n_{Aa}!\frac{1}{2}(n_{a}-n_{Aa})!(2n)!}\]For example, a sample with 2504 individuals, the observed genotypes are 2467/36/1 for AA, Aa and aa. We can do the chi-square test in R.
### Observed genotype counts
n_AA = 2467
n_Aa = 36
n_aa = 1
n = n_AA + n_Aa + n_aa
O_geno = c(n_AA, n_Aa, n_aa)
cat(O_geno)
## 2467 36 1
### Observed allele count and frequency
n_A = n_AA * 2 + n_Aa
n_a = n_aa * 2 + n_Aa
p_A = n_A/(2 * n)
p_a = n_a/(2 * n)
cat(p_A, p_a)
## 0.9924121 0.007587859
### Expected genotype count, under HWE
E_AA = p_A^2 * n
E_Aa = 2 * p_A * p_a * n
E_aa = p_a^2 * n
E_geno = c(E_AA, E_Aa, E_aa)
cat(E_geno)
## 2466.144 37.71166 0.1441693
### Chi-square with df=1 (#genotypes - #alleles)
chisq = sum((O_geno-E_geno)^2/E_geno)
pvalue = 1 - pchisq(chisq, 1)
cat(paste("chisq:", chisq, ";","pvalue: ", pvalue))
## chisq: 5.15844350552391 ; pvalue: 0.02313362043297
We can also use the R package “HardyWeinberg” to do the chi-square test.
### We can also use the R package "HardyWeinberg"
library(HardyWeinberg)
HWChisq(O_geno, cc=0)
## Chi-square test for Hardy-Weinberg equilibrium (autosomal)
## Chi2 = 5.158444 DF = 1 p-value = 0.02313362 D = -0.8558307 f = 0.04538812
It’s hard to do the exact test directly in R because of the factorial calucation. So we also use the R package “HardyWeinberg”. A more common way is to use PLINK “–hwe” option.
HWExact(O_geno)
## Haldane Exact test for Hardy-Weinberg equilibrium (autosomal)
## using SELOME p-value
## sample counts: n = 2467 n = 36 n = 1
## H0: HWE (D==0), H1: D <> 0
## D = -0.8558307 p-value = 0.1318891
We can see that the p-values from chi-square and exact test are quite different. In 2005, Wigginton et al has shown that the chi-square inflated type I error rates. So exact test is a better method.
Linkage disequilibrium
Disequilibrium coefficient (D)
For a pair of biallelic loci, there are four haplotypes. If there is no association between these two SNPs (linkage equilibrium), the haplotype frequencies should be:
- $p_{AB} = p_{A} \times p_{B}$
- $p_{aB} = p_{a} \times p_{B}$
- $p_{Ab} = p_{A} \times p_{b}$
- $p_{ab} = p_{a} \times p_{b}$
However, if the two SNPs have nonrandom association, the LD can be defined by disequilibrium coefficient:
- $D_{AB} = p_{AB} - p_{A} \times p_{B}$
- $P_{AB} = p_{A} \times p_{B} + D_{AB}$
- $p_{aB} = p_{a} \times p_{B} - D_{AB}$
- $p_{Ab} = p_{A} \times p_{b} - D_{AB}$
- $p_{ab} = p_{a} \times p_{b} + D_{AB}$
Other LD measures
More wildely used measures for LD are D’ (LEWONTIN 1964) and $r^{2}$ (HILL and ROBERTSON 1968).
\[D'_{AB} = \frac{D_{AB}}{D_{min}}\] \[D_{min} = max(-p_{A}p_{B}, -p_{a}p_{b}), if D_{AB} < 0\] \[D_{min} = min(p_{A}p_{b}, p_{a}p_{B}), if D_{AB} > 0\]D′ = 1 indicates that at least one of the four possible haplotypes is absent, regardless of the allele frequencies.
\[r^{2} = \frac{D^{2}}{p_{A}p_{a}p_{B}p_{b}}\]$r^{2}$ is a correlation coefficient of 1/0 (all or none) indicator variables indicating the presence of A and B.
Influence Factor
Linkage disequilibrium is influenced by many factors, including,
- selection
- the rate of recombination
- the rate of mutation
- genetic drift
- the system of mating
- population structure
- genetic linkage.
An example in R
Haplotype table
| haplotype | ab | Ab | aB | AB |
|---|---|---|---|---|
| frequency | 0 | 32/5008 | 1/5008 | 4975/5008 |
2*2 allele frequency table
| allele | a | A | Total |
|---|---|---|---|
| b | 0 | 32/5008 | 32/5008 |
| B | 1/5008 | 4975/5008 | 4976/5008 |
| Total | 1/5008 | 5007/5008 | 1 |
#haplotype frequency
pab = 0
pAb = 32/5008
paB = 1/5008
pAB = 4975/5008
cat(pab, pAb, paB, pAB)
## 0 0.006389776 0.0001996805 0.9934105
#allele frequency
pa = paB + pab
pA = pAB + pAb
pb = pab + pAb
pB = paB + pAB
cat(pa, pA, pb, pB)
## 0.0001996805 0.9998003 0.006389776 0.9936102
#D
D = pAB - pA*pB
cat(D)
## -1.275914e-06
#Dmin
Dmin = max(-pA*pB, -pa*pb)
cat(Dmin)
## -1.275914e-06
#D'
Ddot = D/Dmin
cat(Ddot)
## 1
#r-square
rsq = D^2/(pa*pA*pb*pB)
cat(rsq)
## 1.284376e-06
Reference
- ppt from Peter Visscher
- Laird et al, The Fundamentals of Modern Statistical Genetics, 2011
- R package HardyWeinberg
- Lewontin RC. The Interaction of Selection and Linkage. I. General Considerations; Heterotic Models. Genetics, 1964.
- Hill W.G. Linkage disequilibrium in finite populations. Theoretical and Applied Genetics, 1968.
- Gonçalo’s Lecture Notes - Models in Human Genetics