Fastq格式
Fastq是二代测序中常用的原始序列文件格式。
Fastq文件如何得到?
After the sequencing platform generates the sequencing images, the data are analyzed in five steps: image analysis, base calling, bcl conversion, sequence alignment, and variant analysis and counting. CASAVA performs the bcl conversion, sequence alignment, and variant analysis and counting steps, demultiplexes multiplexed samples during the bcl conversion step. Bcl conversion — Converts *.bcl files into *.fastq.gz files (compressed FASTQ files) in CASAVA. Multiplexed samples are demultiplexed during this step.
在测序平台产生测序图像之后,数据分析有5个步骤:图像分析,base calling,bcl文件转换,序列比对和变异分析。illumina的CASAVA软件进行bcl转换,序列比对和变异分析这些步骤。在bcl转换步骤中,多样本数据将会拆分。
Bcl转换 - 用CASAVA软件转换bcl文件为fastq.gz文件(压缩的fastq文件)。多样本数据将会拆分。
示例:
@EAS139:136:FC706VJ:2:5:1000:12850 1:Y:18:ATCACG
AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
+
BBBBCCCC?<A?BC?7@@???????DBBA@@@@A@@
- 第1行:header
- 第2行:序列
- 第3行:固定为“+”
- 第4行:质量值(quality score)
Header
@<instrument>:<run number>:<flowcell ID>:<lane>:<tile>:<x-pos>:<y-pos> <read>:<is filtered>:<control number>:<index sequence>
| Element | Requirements | Description |
|---|---|---|
| @ | @ | Each sequence identifier line starts with @ |
<instrument> |
Characters allowed: a-z, A-Z, 0-9 and underscore | Instrument ID |
<run number> |
Numerical | Run number on instrument |
<flowcell ID> |
Characters allowed: a-z, A-Z, 0-9 | |
<lane> |
Numerical | Lane number |
<tile> |
Numerical | Tile number |
<x_pos> |
Numerical | X coordinate of cluster |
<y_pos> |
Numerical | Y coordinate of cluster |
<read> |
Numerical | Read number. 1 can be single read or read 2 of paired-end |
<is filtered> |
Y or N | Y if the read is filtered, N otherwise |
<control number> |
Numerical | 0 when none of the control bits are on, otherwise it is an even number. |
<index sequence> |
ACTG | Index sequence |
因此@EAS139:136:FC706VJ:2:5:1000:12850 1:Y:18:ATCACG可以解释为:
- 测序仪id为
EAS139 - run number:136
- flowcell ID:FC706VJ
- lane:2
- tile:5
- x_pos:1000
- y_pos:12850
- read:1,代表是单端测序
- is filtered:Y,代表是filtered
- control number:18
- index sequence:ATCACG
Phred quality score
A Phred quality score is a measure of the quality of the identification of the nucleobases generated by automated DNA sequencing.[1][2] It was originally developed for Phred base calling to help in the automation of DNA sequencing in the Human Genome Project.
Phred quality score是用来测定DNA自动测序中每个核算的测序质量的。它最早是为了人类基因组计划中的程序Phred base calling而开发的。
- 公式
Q = -10 * log10(P) <==> P = 10 ^(-Q/10)
这里Q为Phred quality score,P为base-calling的error probabilities(错误率)。
- 具体表格
| Phred Quality Score | Probability of incorrect base call (错误率) | Base call accuracy(准确率) |
|---|---|---|
| 10 | 十分之一(1 in 10) | 90% |
| 20 | 百分之一(1 in 100) | 99% |
| 30 | 千分之一(1 in 1000) | 99.9% |
| 40 | 万分之一(1 in 10,000) | 99.99% |
| 50 | 十万分之一(1 in 100,000 ) | 99.999% |
| 60 | 百万分之一(1 in 1,000,000) | 99.9999% |
Encoding of Quality score
| 类型 | Quality score范围 | ASCII范围 |
|---|---|---|
| Sanger format | 0 ~ 93(rarely exceeds 60) | 33 ~ 126(Phred+33) |
| Solexa/Illumina 1.0 format | -5 ~ 40 | 59 ~ 104(Solexa+64) |
| Illumina 1.3+ | 0 ~ 40 | 64 ~ 104(Phred+64) |
| Illumina 1.5+ | 3 ~ 40 | 67 ~ 104(Phred+64) |
| Illumina 1.8+ | 0 ~ 41 | 33 ~ 74(Phred+33) |
For raw reads, the range of scores will depend on the technology and the base caller used, but will typically be up to 41 for recent Illumina chemistry
对于原始的序列,质量值的范围取决于所用的技术和base caller,但是对于目前的Illumina技术,最多能达到41。
另外,常用软件FastQC和Trimmomatic均可以自动判断encoding的。
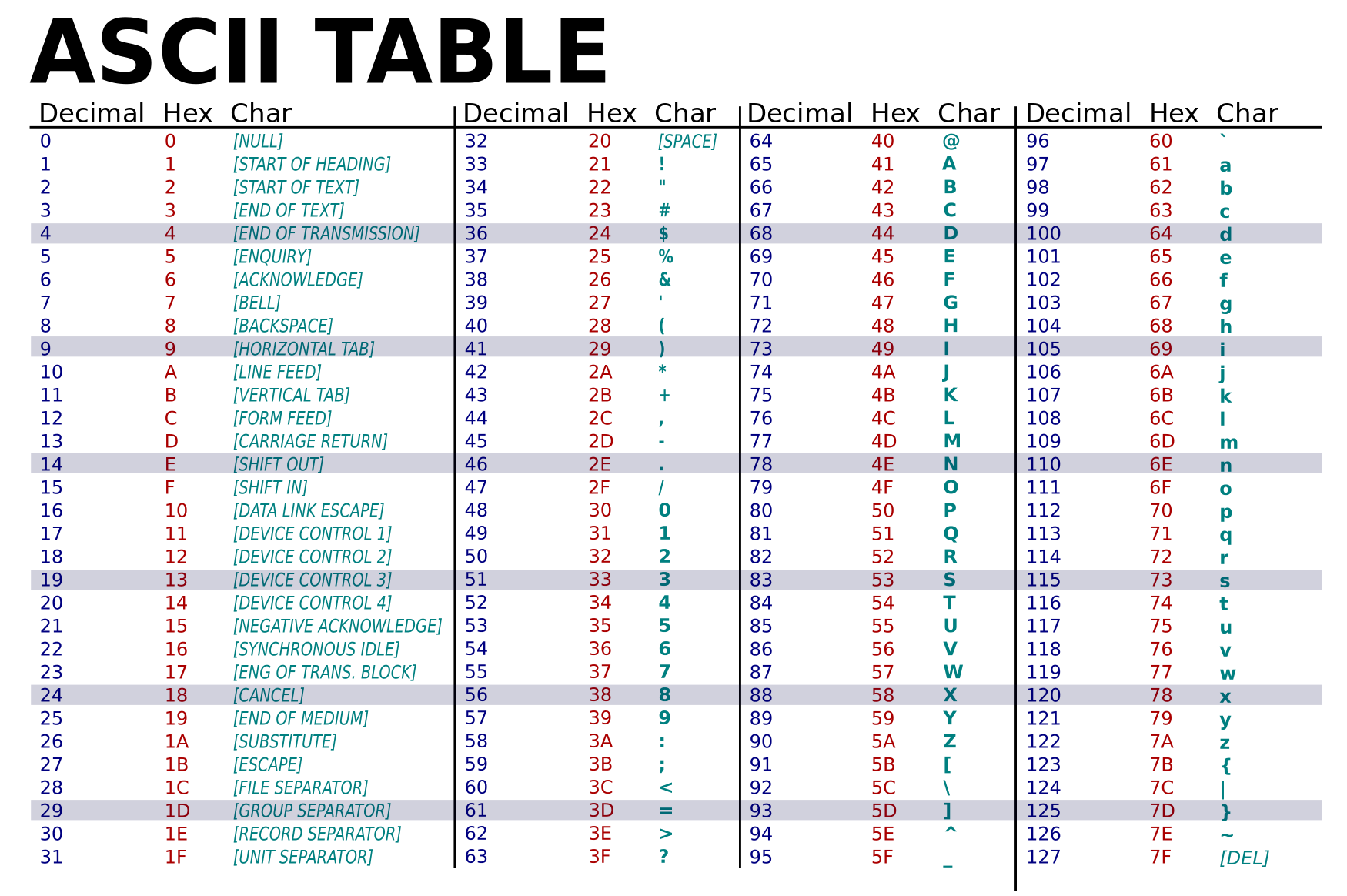
因此,质量值BBBBCCCC?<A?BC?7@@???????DBBA@@@@A@@:
- 若采用Phred+64编码,应该为
2,2,2,6,6,6,6,6,6,6,6,6,6,9,9,9,9,9,9,9,9,9,9,9,9,9,9,2,6,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,9,6,6,6,6,6,6,6,6,6,6,2,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,2,6,6,6,6,6,6,6 - 若采用Phred+32编码,应该为
33,33,33,37,37,37,37,37,37,37,37,37,37,40,40,40,40,40,40,40,40,40,40,40,40,40,40,33,37,40,40,40,40,40,40,40,40,40,40,40,40,40,40,40,40,40,40,40,40,40,40,40,40,40,40,37,37,37,37,37,37,37,37,37,37,33,37,37,37,37,37,37,37,37,37,37,37,37,37,37,37,37,37,37,37,37,37,37,37,37,37,37,33,37,37,37,37,37,37,37
故,很明显,使用的是“Phred+32”编码,且质量值均大于30,错误率均小于千分之一,准确率大于99.9%。
ASCII
ASCII(American Standand Code for Information Interchange),是一套计算机编码系统。