中国新冠死亡率
更新:
根据国家卫健委的统计数据,
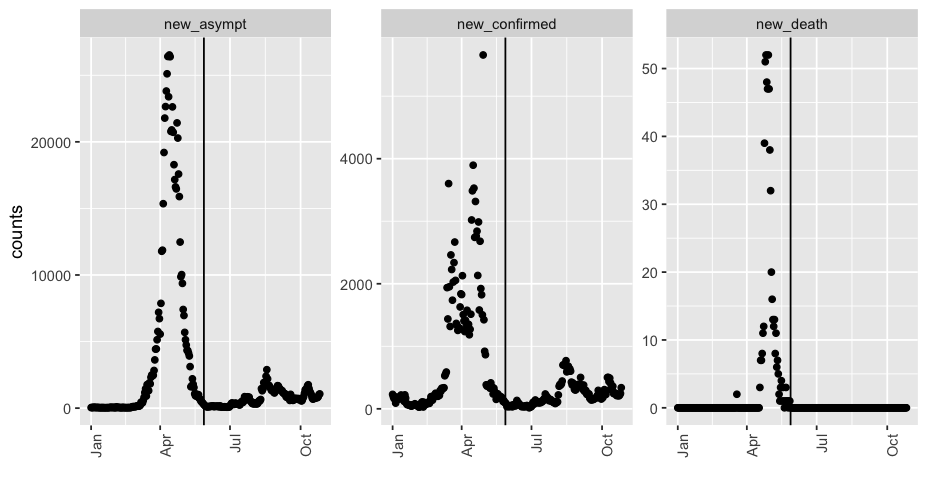
- 从2022年1月1日到2022年10月26日,国内共有新增确诊病例累计156087,新增无症状感染者累计785801,新增死亡累计590。死亡率仅为0.0006264014(万分之六)
- 从2022年5月28日算起,国内新增死亡病例为0,但却累计新增了确诊病例34331,无症状感染者111841。
不知道我的计算是否有误,代码见下方。
data = readLines("china_cdc.txt")
days = data[grepl("2022$",data) & !grepl("National Health Commission Update", data)]
new_confirmed = as.integer(sapply(strsplit(data[grep("Confirmed:", data)], "Confirmed: | new"), function(x) sub(",", "",x[2])))
new_asympt = as.integer(sapply(strsplit(data[grep("Asymptomatic:", data)], "Asymptomatic: | new"), function(x) sub(",", "",x[2])))
new_death = as.integer(sapply(strsplit(data[grep("Deaths:", data)], "Deaths: | new"), function(x) sub(",", "",x[3])))
res = data.frame(days=as.Date(days, "%b %d, %Y"), new_confirmed, new_asympt, new_death)
library(ggplot2); library(tidyverse)
ggplot(res %>% pivot_longer(-days), aes(x=days, y=value)) +
geom_point() + xlab("")+ ylab("counts")+
facet_wrap(vars(name), scales="free_y") + theme(axis.text.x = element_text(angle = 90, hjust = 1)) + geom_vline(xintercept=as.Date("2022-05-28"))
res %>% filter(days >= as.Date("2022-05-28")) %>% summarise(sum(new_confirmed), sum(new_asympt), sum(new_death), n())
### sum(new_confirmed) sum(new_asympt) sum(new_death) n()
### 1 34331 111841 0 152
res %>% summarise(sum(new_confirmed), sum(new_asympt), sum(new_death), n())
### sum(new_confirmed) sum(new_asympt) sum(new_death) n()
### 1 156087 785801 590 299
590/(156087+785801)
### [1] 0.0006264014